


      
|

|
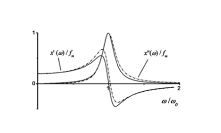
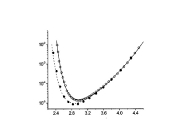
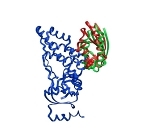
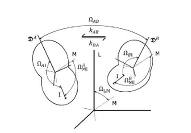
Protein Structure ElucidationProtein structure elucidation comprises a wide spectrum of computational methods targeted to provide three dimensional structures of proteins using combinations of model inter-atomic force fields, database pseudo potentials, and experimental data, which distinguishes it from folding paradigm claiming not to use experimental data at all. My particular interest in this field was originated from my works on protein dynamics when I learned that mobility of proteins often closely related to their structures. If so, the experimental information about protein mobility could be used to solve protein structures. I started to work on these problems at University of Maryland in the Lab of Professor David Fushman and continued at NIH collaborating with Charles Schwieters and Marius Clore. Our first work on this subject (JACS 2007) was a "prove of principle" where we demonstrated that the information about rotational tumbling of proteins in solution, which is available from NMR relaxation data for example, can be used to restrain overall shape of proteins. A more advanced version of the same approach was implemented into Xplor-NIH structure elucidation platform (JACS 2009, JACS 2010). Together with the ability to characterize long-range global parameters the protein dynamics also possess more detailed information, like the information about residue specific NH orientations. The most recent development of this method (JACS 2011) uses this residue specific orientation information together with the overall shape restrains for structure determination of globular proteins. Current implementation of the method can be also used for data driven docking of protein complexes. This implementation has a number of useful features like "on-the-fly" fitting of optimal experimental temperature settings and filtering for random data outliers. Figure on the right presents the results of structure calculations of EIN protein domain with NMR relaxation rates as experimental input performed at different temperature setting. Related articles: |