



      
|

|
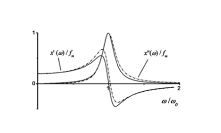
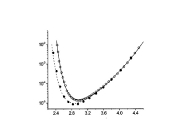
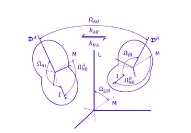
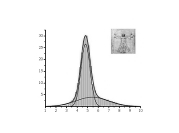
Protein DynamicsI started to work on these problems since I accepted a postdoctoral position at University of Maryland in the Lab of Professor David Fushman and continued these researches at NIH collaborating with Charles Schwieters and Marius Clore. I am trying to understand the relationships between internal dynamics and structure of proteins with the purpose to develop methods to extract information about protein internal mobility from experimental data. The key for both problems is theoretical modeling of protein internal motions. I developed a model which treats the relationships between large scale reorientations of protein domains and overall rotational diffusion tumbling. The cartoon picture on the right is from our publication which discusses coupling between overall diffusion tumbling and internal mobility of proteins (JCP 2012). This theoretical work establishes a closed form solution for a quite general problem of interconversion between a set of discrete conformation states. When working on this paper I was really lucky to have a possibility to discuss my progress with Attila Szabo and Alex Berezhkovkii who helped me a lot in formulation of these results. We hope that we will be able to implement this model into standard protocols of protein structure elucidation. This should provide the possibility to modify these protocols in such a way that it would be possible not only to define protein structure but also to derive parameters of it's internal motions. An integral part of this proposal is a suitable method for computation of protein diffusion tensor. Earlier I developed such a method (JACS 2006) based on fast sampling of protein surface and then converting topology of this surface into equivalent ellipsoid shape which, in turn, is used for evaluation of protein rotation diffusion tensor in closed forms. Four bottom figures on the right illustrate essential steps of this method, which is sufficiently fast to be implemented into molecular dynamics protocols. The initial package, ELM, implementing this method was a strange combination of original C based SURF program, written by Amitabh Varshney in 1994, which was necessary to map protein surface, and an in-house MATLAB wrapper, that was doing all the job calculating protein diffusion tensor. This package is still freely available upon request. But a more advanced version of the same method is implemented in C++ core of Xplor-NIH. This is the method which we employed before for protein structure elucidation and this is the method that we are going to utilize for this project. Related articles:Y. Ryabov, G. Marius Clore, Charles D. Schwieters "Coupling between internal dynamics and rotational diffusion in the presence of exchange between discrete molecular conformations", J. Chem. Phys. 136: # 034108 (2012) with Supplementary Y. Ryabov, C. Geraghty, A. Varshney, and D. Fushman, "An Efficient Computational Method for Predicting Rotational Diffusion Tensors of Globular Proteins Using an Ellipsoid Representation", J. Am. Chem. Soc. 128: 15432-15444 (2006) with Supporting information |