



      
|

|
Genome Evolution
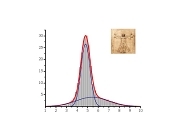
This work branched from my protein dynamics activity by a quite unusual way. When we have got in our hands a fast method for protein diffusion tensor predictions we run it over a set of protein structures representing more than 800 different folds. We described these results in a paper about this method. However one aspect was puzzling: statistical distribution of overall rotation correlation time seemed to obey a lognormal law. This fact was not really related to the subject of that work and we just briefly mentioned it in supporting information file. But why it was lognormal? I asked many people about possible reasons for this lognormality and got a wide spectrum of different answers ranging from "one should be not worrying about this" to "this is selection bias of this set of protein folds". Until, once I got a real lead when I was discussing these observations with Professor Alexei Petrovich Sokolov at Akron University in late 2005. He mentioned that a lognormal distribution usually originates from Kolmororov multiplicative process. This was the real moment of conception for this project. The application of Kolmogorov model to our observations lead me from statistics of protein rotation diffusion correlation times to genomics. It took some time and a great deal of investigations, thinking and rationalizing. Most of this work I performed at Purdue University were I consulted a lot with Proffesor Michael Gribskov. Finally we arrived to analyzing distribution of exon lengths of different genomes. Figures on the rigth summarize the results of this work. We observed that all genomes show two lognormal peaks, one of which can be strongly correlated with the exons involved in alternative splicing. Kolmogorov model provides formal background for understanding lognormal nature of these peaks. It also suggests that the parameters of lognormal peaks observed in exon seize distributions can serve as a measures of the amount of changes in a genome during its evolution. Beside this, Kolmogorov model implies that process of intron insertion should be independent of exon size. This implication contrasts with usual opinion that larger exons are bigger targets for transposons and therefore should be split more frequently. Related article:Y. Ryabov and M. Gribskov, "Spontaneous symmetry breaking in genome evolution", Nucleic Acids Research 36: 2756-2763 (2008) with Supplementary data. |